- HOME
- Enzyme List
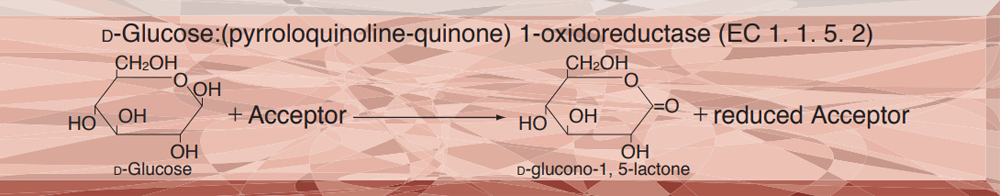
- GLD-321 GLUCOSE DEHYDROGENASE (PQQ-dependent)
GLD-321
GLUCOSE DEHYDROGENASE (PQQ-dependent) from Microorganism
PREPARATION and SPECIFICATION
| Appearance | Purple amorphous powder, lyophilized | |
|---|---|---|
| Activity | GradeⅢ 500 U/mg-solid or more | |
| Contaminants | Glucose dehydrogenase (NAD-dependent) |
≤ 1.0×10-3 % |
| Hexokinase | ≤ 1.0×10-3 % | |
| Stabilizers | Ca2+, BSA | |
PROPERTIES
| Stability | Stable at − 20℃ for at least one year (Fig.1) |
|---|---|
| Molecular weight | approx. 100,000 (by gel filtration) |
| Michaelis constant | 4.8 mM (D-Glucose) |
| Inhibitors | Cu2+, Pb2+, Ag+ |
| Optimum pH | 7.0 (Fig.2) |
| Optimum temperature | 37 ℃ (Fig.3) |
| pH Stability | pH 3.5−8.5 (25 ℃, 16 hr) (Fig.4) |
| Thermal stability | below 50 ℃ (pH 7.5, 30 min) (Fig.5) |
| Substrate specificity | (Table 1) |
| Effect of various chemicals | (Table 2) |
APPLICATIONS
This enzyme is useful for enzymatic determination of D-glucose.
ASSAY
Principle
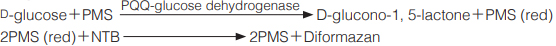
The formation of diformazan formed by the reduction of nitrotetrazorium blue (NTB) with phenazine methosulfate (PMS), which is red, is measured at 570 nm by spectrophotometry.
Unit definition
One unit causes the formation of one half micromole of diformazan per minute under the conditions detailed below.
Method
Reagents
| A. D-Glucose solution | 1 M: 1.8 g of D-glucose (MW = 180.16) / 10 mL of H2O (keep this solution at room temperature for at least 3 hours before use). |
|---|---|
| B. PIPES-NaOH buffer, pH 6.5 | 50 mM: Weigh out 1.51 g of PIPES (MW = 302.36), suspend in 60 mL of H2O, dissolve with 5 N NaOH, and add 2.2 mL of 10 % Triton X-100. After adjusting pH to 6.5 ± 0.05 at 25℃ with 5 N NaOH, make up to 100mL with H2O. |
| C. PMS solution | 3.0 mM: 9.19 mg of phenazine methosulfate (MW = 306.34) / 10 mL of H2O |
| D. NTB solution | 6.6 mM: 53.96 mg of NTB (MW = 817.65) / 10 mL of H2O |
| E. Enzyme diluent | 50 mM PIPES-NaOH buffer, pH 6.5, containing 1 mM CaCl2, 0.1 % Triton X-100, 0.1 % BSA |
Procedure
1.Prepare the following reaction mixture in a brownish bottle shortly before use, and store on ice.
| 0.9 mL | D-glucose solution | (A) |
| 25.5 mL | PIPES-NaOH buffer, pH 6.5 | (B) |
| 2.0 mL | PMS solution | (C) |
| 1.0 mL | NTB solution | (D) |
| Concentration in assay mixture | |
|---|---|
| PIPES-buffer | 42 mM |
| D-glucose | 30 mM |
| PMS | 0.20 mM |
| NTB | 0.22 mM |
2.Pipette 3.0 mL of working solution into a plastic test tube and equilibrate at 37 ℃ for approximately 5 minutes.
3.Add 0.1 mL of enzyme solution* and mix by gentle inversion.
4.Record the increase of optical density at 570 nm against water for 4 to 5 minutes with a spectrophotometer thermostated at 37 ℃, and calculate the ΔOD per minute from the initial linear portion of the curve (ΔOD test).
At the same time, measure the blank rate (ΔOD blank) by the same method as in the test except that the enzyme diluent (E) is added instead of the enzyme solution.
*Dissolve the enzyme preparation on ice cold enzyme diluent (E) and dilute to 0.1−0.8 U/mL with the same buffer, immediately before the assay. (A plastic tube is recommended because of viscous properties of the liquid.)
Calculation
Activity can be calculated by using the following formula :
Volume activity (U/mL) =
-
ΔOD/min (ΔOD test−ΔOD blank)×Vt×df
20.1×1.0×Vs
= ΔOD×1.54×df
Weight activity (U/mg) = (U/mL)×1/C
| Vt | : Total volume (3.1 mL) |
| Vs | : Sample volume (0.1 mL) |
| 20.1 | : Half a millimolar extinction coefficient of diformazan (cm2/0.5 micromole) |
| 1.0 | : Light path length (cm) |
| df | : Dilution factor |
| C | : Enzyme concentration in dissolution (c mg/mL) |
REFERENCES
1)K.Matsushita et al.; FEMS Microbiology Letters, 55, 53 (1988).
Table 1. Substrate Specificity of PQQ-Glucose dehydrogenase
-
Substrate (50mM) Relative activity(%) D-Glucose 100.0 L-Glucose 0.3 D-Xylose 15.0 2-Deoxy-glucose 4.9 L-Sorbose 0.5 D-Mannose 10.8 D-Fructose 0.3 -
Substrate (50mM) Relative activity(%) Galactose 16.0 D-Lactose 68.9 D-Sorbitole 0.2 D-Mannitol 0.0 Sucrose 0.2 Inositol 0.0 Maltose 107.0
Table 2. Effect of Various Chemicals on PQQ-Glucose dehydrogenase
[The enzyme dissolved in 50mM PIPES-NaOH buffer, pH 6.5 contg. 1mM CaCl2, 0.1% Triton X-100 (5U/mL) was incubated with each chemical at 25℃ for 1hr.]
-
Chemical Concn.(mM) Residual
activity(%)None - 100 Metal salt 2.0 MgSO4 108 CaCl2 108 Ba(OAc)2 105 FeCl3 79 CoCl2 42 MnCl2 105 ZnCl2 45 Cd(OAc)2 107 NiCl2 101 CuSO4 0 Pb(OAc)2 0 AgNO3 0 HgCl2 77 2-Mercaptoethanol 2.0 99 PCMB 1.0 97 -
Chemical Concn.(mM) Residual
activity(%)MIA 2.0 87 NEM 2.0 100 IAA 2.0 98 Hydroxylamine 2.0 19 EDTA 5.0 79 O-Phenanthroline 2.0 7 α,α′-Dipyridyl 1.0 103 Borate 5.0 110 NAF 2.0 111 NaN3 2.0 115 Triton X-100 0.10 % 101 Brij 35 0.10 % 22 Tween 20 0.10 % 104 Span 20 0.10 % 60 Na-Cholate 0.10 % 67 SDS 0.05 % 33 DAC 0.05 % 113
Ac, CH3CO; PCMB, p-Chloromercuribenzoate; MIA, Monoiodoacetate; NEM, N-Ethylmaleimide; IAA, Iodoacetamide; EDTA, Ethylenediaminetetraacetate; SDS, Sodium dodecyl sulfate; DAC, Dimethylbenzylalkylammonium chloride.
-
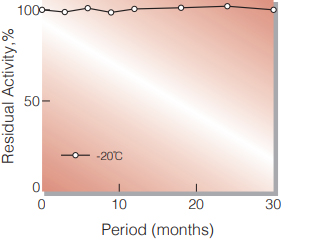
Fig.1. Stability (Powder form)
(kept under dry conditions)
-
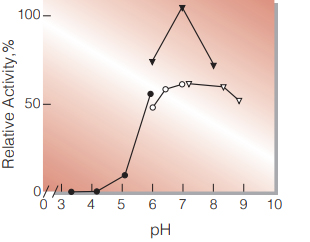
Fig.2. pH-Activity
25℃, in 50mM buffer solution; ●̶●,acetate; ▼̶▼, phosphate; ○̶○, PIPES; ▽̶▽, Tris-HCl.
-
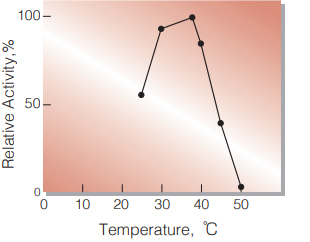
Fig.3. Temperature Activity
(in 42mM PIPES-NaOH buffer, pH 6.5)
-
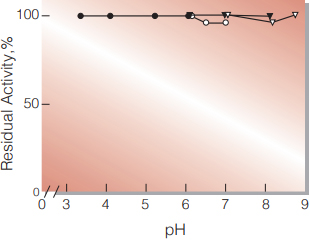
Fig.4. pH-Stability
25℃, 16 hr-treatment with 50mM buffer solution contg. 1mM CaCl2; ●̶●,acetate; ▼̶▼, phosphate; ○̶○, PIPES; ▽̶▽, Tris-HCl.
-
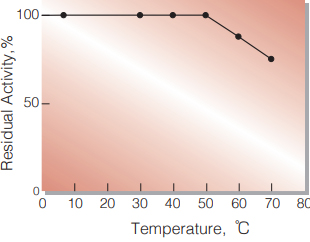
Fig.5. Thermal stability
30min.-treatment with 50mM PIPES-NaOH buffer, pH 6.5 contg. 1mM CaCl2 enzyme concentration: 5.0 U/mL
活性測定法(Japanese)
1. 原理
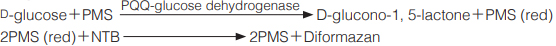
PMS(phenazine methosulfate)を介してNTB (nitrotetrazorium blue)を還元し,生成したdiformazan の570nmにおける吸光度を測定する。
2.定義
下記条件下で1分間あたり1/2マイクロモルの diformazanを生成する酵素量を1単位(U)とする。
3.試薬
- 1M D-グルコース溶液(1.8gのD-グルコースを蒸留水に溶解し,10 mLとし,室温で3時間経過したものを使用する)
- 50mM PIPES-NaOH緩衝液,pH6.5(1.51gの PIPESを60 mLの蒸留水に懸濁し,5N NaOHで溶解後,10%のTritonX-100溶液2.2 mLを加える。5N NaOHを使って25℃でpHを6.5±0.05 に調整後,蒸留水で100 mLとする)
- 3mM PMS溶液(9.19mgのPMSを10 mLの蒸留水に溶解する)
- 6.6mM NTB溶液(53.96mgのNTBを10 mLの蒸留水に溶解する)
酵素溶液:酵素標品を予め氷冷した1mM CaCl2と 0.1% TrironX-100と0.1%BSAを含む 50mM PIPES-NaOH緩衝液,pH6.5で 0.1~0.8U/mLに希釈する。
4.手順
1.褐色瓶中に下記反応混液を調製し,氷冷保存する。
| 0.9 mL | D-グルコース溶液 | (A) |
| 25.5 mL | PIPES-NaOH緩衝液 | (B) |
| 2.0 mL | PMS溶液 | (C) |
| 1.0 mL | NTB溶液 | (D) |
2.反応混液3.0 mLをプラスチック製のキュベット(d= 1.0cm)に分注し37℃で約5分間予備加温する。
3.酵素溶液を0.1 mLを添加し,緩やかに混和後,水を対照 に37℃に制御された分光光度計で570nmの吸光度変化を4~5分間記録し,その直線部分から1分間あたりの吸光度変化を求める(ΔODtest)。
4.盲検は反応混液①に酵素液の代りに酵素希釈液 (1mM CaCl2と0.1% TritonX-100と0.1% BSAを含む50mM PIPES-NaOH緩衝液,pH6.5)0.1 mLを加え,上記同様に操作を行い,1分間当たりの吸光度変化を求める(ΔODblank)。
5.計算式
-
U/mL =
-
ΔOD/min (ΔOD test−ΔOD blank)×3.1×df
20.1×1.0×0.1
| = ΔOD/min×1.54×df | |
| U/mg | =U/mL×1/C |
| 20.1 | : diformazanの1/2ミリモル分子吸光係数 (cm2/0.5 micromole) |
| 1.0 | : 光路長(cm) |
| C | : 溶解時の酵素濃度(C mg/mL) |
CONTACT
-
For inquiries and cosultations regarding our products, please contact us through this number.
- HEAD OFFICE+81-6-6348-3843
- Inquiry / Opinion
