- HOME
- Cell lysis & cDNA Synthesis kit for real-time PCR (For cultured cells) SuperPrepTMII Cell Lysis & RT Kit for qPCR/
SuperPrepTMII Cell Lysis Kit for qPCR
-
Cell lysis & cDNA Synthesis kit for real-time PCR (For cultured cells)
SuperPrepTMII Cell Lysis & RT Kit for qPCR/
SuperPrepTMII Cell Lysis Kit for qPCRCode No. SCQ-401 100 reactions/Code No. SCQ-501 100 reactions
DESCRIPTION
-
-
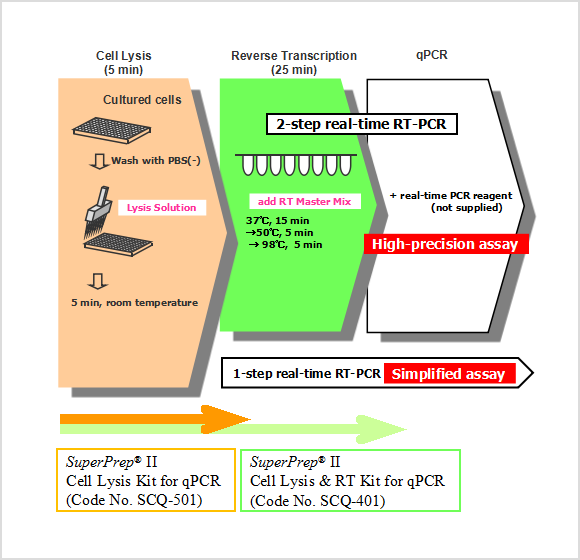
SuperPrep™ II Cell Lysis & RT Kit for qPCR (Code No. SCQ-401) consists of "Lysis Reagents" and "RT Reagents" for synthesis of cDNA templates for real-time PCR assays.
"Lysis Reagents" prepares cell lysates containing RNAs that can be used as templates for reverse transcription. "RT Reagents" contains reagents for reverse transcription, optimized for efficient cDNA synthesis from crude lysates. The synthesized cDNA can be applied to real-time PCR directly. This assay system is suitable for high-throughput assays. In the previous version, it was necessary to add Stop Solution after adding Lysis Solution. In SuperPrep™ II, cell lysates with lysis solution can be used as templates for cDNA synthesis directly. Moreover, high sensitivity detection is possible from a wide variety of mammalian cells than in the previous version. With this assay system, it is possible to synthesize template cDNA for real-time PCR from cultured cells conveniently and quickly.
SuperPrep™ II Cell Lysis Kit for qPCR (Code No. SCQ-501) is an option of "Lysis Reagents". The cell lysate prepared by "Lysis Reagents" can be applied to one-step real-time PCR.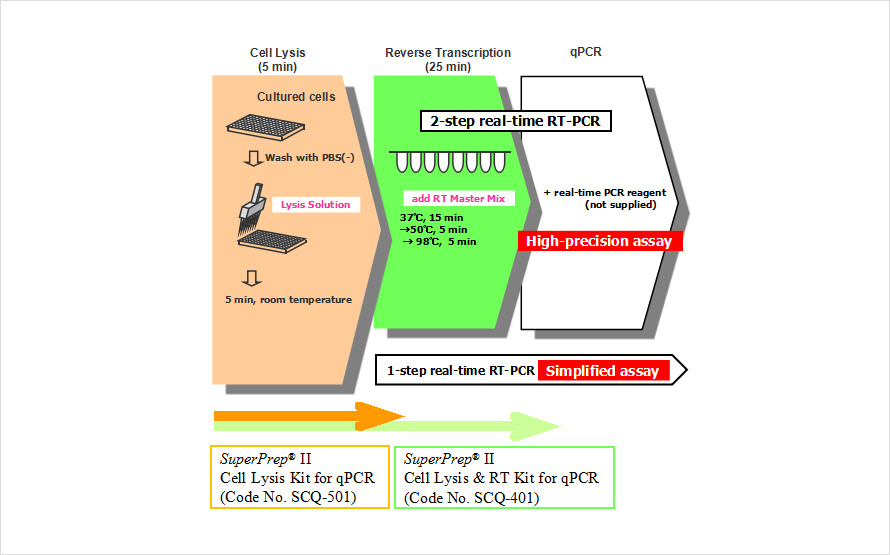
-
Features
- ・ RNA purification is not necessary.
- ・ High-quality cDNA can be obtained from cell lysates.
- ・ Reduction of dispersion on high-throughput assay.
- ・ Various real-time PCR regents can be applied.
Details
Applications
DNA synthesis for real-time PCR from mammalian cultured cells(Code No. SCQ-401)
Total RNA preparation for one-step real-time PCR from mammalian cultured cells (Code No. SCQ-501)
Storage condition
Store at -20ºC
Components
The reagent includes the following components for 20 reactions (SCQ-401S) or 100 reactions (SCQ-401, SCQ-501).
SCQ-401S and SCQ-401 contain two separate packages named "Lysis Reagents" and "RT Reagents", respectively. All reagents should be stored at -20°C.
Table 1.SuperPrep™ II Cell Lysis & RT Kit for qPCR (Code No.SCQ-401, SCQ-401S)
| <Lysis Reagents> | SCQ-401 | SCQ-401S (SAMPLE) |
|---|---|---|
| Lysis Solution | 6.5 mL | 1.3 mL |
| gDNA Remover | 33 µL | 6.6 µL |
| RNase Inhibitor | 110 µL | 22 µL |
| <RT Reagents> | SCQ-401 | SCQ-401S (SAMPLE) |
|---|---|---|
| 5 × RT Master Mix | 860 µL | 172 µL |
| 5 × RT Master Mix no-RT Control | 86 µL | 17 µL |
| Nuclease-free Water | 1.7 mL × 2 | 680 µL |
Table 2.SuperPrep™ II Cell Lysis Kit for qPCR (Code No.SCQ-501)
| Lysis Solution | 6.5 mL |
|---|---|
| gDNA Remover | 33 µL |
| RNase Inhibitor | 110 µL |
Application Data
Example 1.High-quality cDNA can be obtained from several kind of cell lysates.
The optimized lysis solution efficiently inhibits RNA degradation during treatment. RNA in the lysate is stable on ice for at least 6 h. High-quality cDNA can be synthesized using highly efficient reverse transcriptase "ReverTra Ace™" with low contamination of genomic DNA because of preceding DNase I treatment. The reverse transcriptase is supplied as a master mix reagent containing optimally mixed primers (random and oligo dT) to achieve effective cDNA synthesis.
Table 1 Cells tested by this system
| Call Name | Adherent Non-adherent |
Species | Remarks | |
|---|---|---|---|---|
| 1 | HPA | Adherent | H.sapiens | preadipocytes (primary cell) |
| 2 | HEK | Adherent | H.sapiens | epidermal keratinocytes (primary cell) |
| 3 | HA | Adherent | H.sapiens | astrocytes (primary cell) |
| 4 | HDF | Adherent | C. griseus | dermal fibroblasts (primary cell) |
| 5 | HBEpC | Adherent | H.sapiens | bronchial epithelial cells (primary cell) |
| 6 | HUVEC | Adherent | H.sapiens | umbilical vein endothelial cells (primary cell) |
| 7 | HPAEC | Adherent | H.sapiens | pulmonary artery endothelial cells (primary cell) |
| 8 | HC | Adherent | H.sapiens | chondrocytes (primary cell) |
| 9 | HOb | Adherent | H.sapiens | osteoblasts (primary cell) |
| 10 | HSkMC | Adherent | H.sapiens | skeletal muscle cells (primary cell) |
| 11 | HAOSMC | Adherent | H.sapiens | Aortic smooth muscle cells (primary cell) |
| 12 | HFDPC | Adherent | H.sapiens | hair follicle dermal papilla Cells (primary cell) |
| 13 | HeLa S3 | Adherent | H.sapiens | cervix carcinoma cell line |
| 14 | HepG2 | Adherent | H.sapiens | hepatocellular carcinoma cell line |
| 15 | Jurkat | Non-adherent | H.sapiens | T lymphocyte cell line |
| 16 | K562 | Non-adherent | H.sapiens | myelogenous leukemia cell line |
| 17 | THP-1 | Non-adherent | H.sapiens | acute monocytic leukemia cell line |
| 18 | U937 | Non-adherent | H.sapiens | leukemic monocyte lymphoma cell line |
| 19 | HMNC | Non-adherent | H.sapiens | Mononuclar cells (primary cell) |
Example 2.Stability test of the cell lysates.
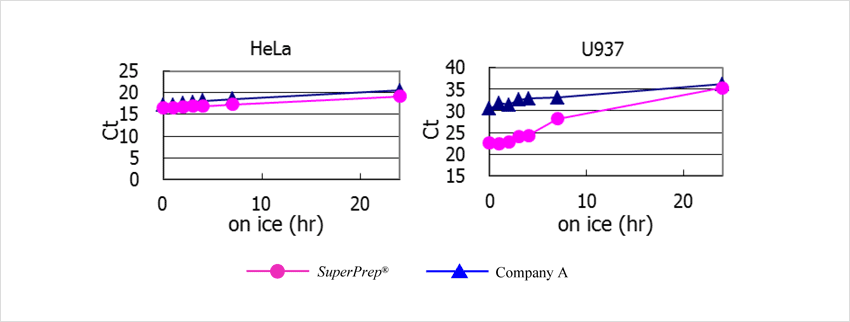
cDNA were synthesized from lysates that had been left on ice for 0–24 h after lysing of 4×104 HeLa and U937 cells using SuperPrep™. β-actin genes were detected using TaqMan™ real-time PCR assay with THUNDERBIRD™ Probe qPCR Mix (Code No. QPS-101). The results were compared with that from the other company’s system (Company A). The results suggest that the RNA in the cell lysates is stable for at least 2 h. Lysates from U937 cells showed higher RNase activity than the other cells and tended to deteriorate in storage over 2 h.
NOTE:
RNase activity depends on the type and number of cells. The cell lysates should be placed on ice after preparation and cDNA should be synthesized immediately after preparing the lysates to minimize RNA degradation.
Example 3.Various real-time PCR reagents can be applied.
The synthesis cDNA can be used in various real-time PCR assay (TaqMan™ probe, SYBR® Green etc.).
In addition, the cell lysate can be applied to one-step real-time PCR reagents.
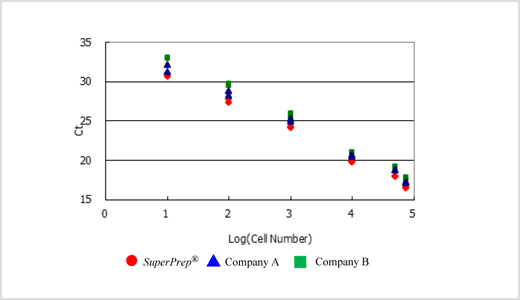
TaqMan™ Probe assay using cDNA synthesized from cell lysates prepared by SuperPrep™
cDNAs were synthesized using the cell lysates (8 µL) prepared from 7.5x104, 5x104, 1x104, 1x103, 1x102 and 1x10] HeLa S3 cells by SuperPrep™ in 40 µL reaction. β-actin genes were detected by various real-time PCR reagents with TaqMan™ real-time PCR assay. Successful amplifications were obtained from all reagents tested and the THUNDERBIRD™ Probe qPCR Mix tended to show a better Ct than the other tested methods.
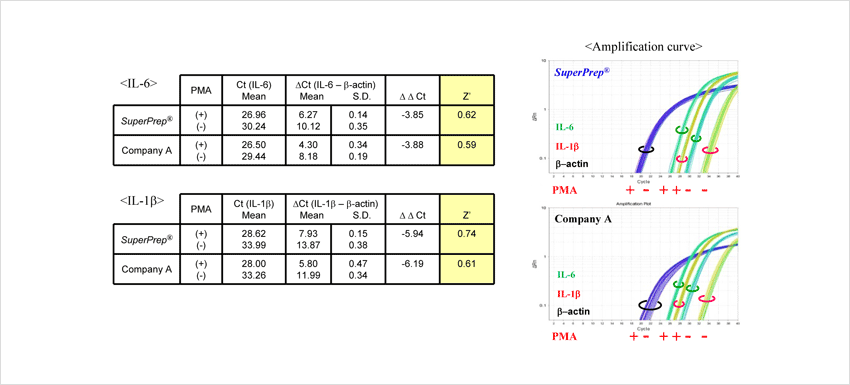
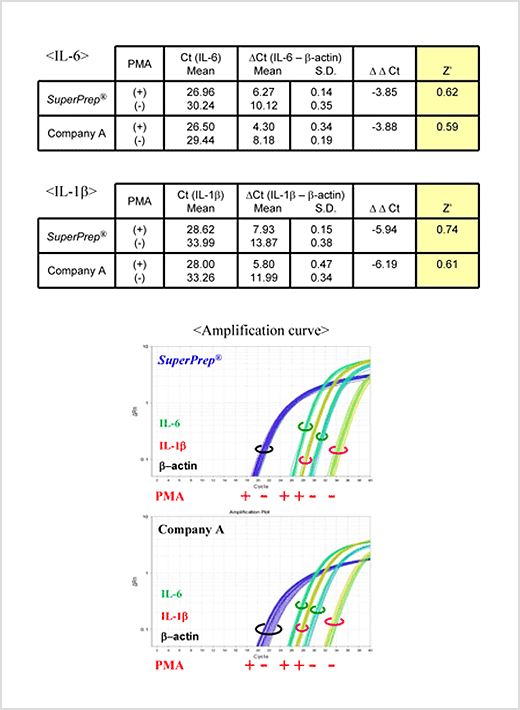
Example 4.Evaluation of the assay variation
HeLa S3 cells were incubated with or without 100 nM phorbol 12-myristate 13-acetate (PMA) for 24 h after seeding at 2 ×104 cells/well in a 96-well culture plate. cDNA were synthesized from the lysates prepared from the cells washed with PBS(-). IL-6, IL-1β and β-actin genes were detected by TaqMan™ real-time PCR assay with THUNDERBIRD™ Probe qPCR Mix (Code No. QPS-101). After compensation of the Cts of IL-6 and IL-1β by that of β-actin, the ΔΔCt between with or without PMA and Z’ factors* were calculated.
Z’ factors from SuperPrep™ were superior to that from the other system (Company A).
*The Z’ factor is a simple statistical parameter that is used to assess the quality of high-throughput screening (HTS) assays. A Z’ score of ≥0.5 is generally considered to indicate good quality Z’ can be calculated by the following formula.
Z’= 1-3 x [Δ Ct(+) standard deviation + Δ Ct(-) standard deviation]/| Δ Δ Ct|