- HOME
- QuantAccuracyTM, RT-RamDATM cDNA Synthesis Kit
-
QuantAccuracyTM, RT-RamDATM cDNA Synthesis Kit
Code No. RMQ-101T 24 reactions, RMQ-101 96 reactions
DESCRIPTION
-
-
The QuantAccuracy™, RT-RamDA™ cDNA Synthesis Kit (Code No. RMQ-101) is an efficient and convenient kit to synthesize cDNA from single cells or trace amounts of RNA for real-time PCR analysis.
By using this kit, cDNA covering full-length total RNA can be synthesized, and gene expression analysis can be performed with high sensitivity.
This kit uses reverse transcription with random displacement amplification (RT-RamDA™) (see method in reference (1)). RT-RamDA™ is a novel cDNA amplification method that utilizes the strand displacement activity of reverse transcriptase, which can detect not only poly(A) RNA but also non-poly(A) RNA with high sensitivity. For this reason, the RT-RamDA™ method is characterized by its ability to detect more genes than conventional technology.
*RT-RamDA™ and RamDA-seq™are trademarks of RIKEN, Institute of Physical and Chemical Research, Japan
Reference (1) Tetsutaro Hayashi*, Haruka Ozaki*, Yohei Sasagawa, Mana Umeda, Hiroki Danno and Itoshi Nikaido. Single-cell full-length total RNA sequencing uncovers dynamics of recursive splicing and enhancer RNAs. Nature Communications. 2018.
-
Features
- - cDNA can be prepared from a single cell or a small amounts of input RNA
Between 1–500 cells or 10 pg–10 ng total RNA should be used. - - Low-copy genes can be analyzed
By amplifying cDNA to tens of times from RNA, low-copy genes can be analyzed from samples containing small amounts of RNA. - - Very accurate quantification
Amplification occurs randomly in RT-RamDA™, which can reduce amplification bias. - - Minimize input samples
Because amplified cDNA can be obtained, valuable samples are saved. - - Can be used with a variety of real-time PCR reagents
This kit can be used in combination with a variety of real-time PCR reagents and is compatible with both SYBR® Green I and TaqMan® assays.
THUNDERBIRD™ qPCR Mix (Code No. QPS-101, QPS-201, QPX-201), and KOD SYBR® qPCR Mix (Code No. QKD-201) can be used.

This is the example of preparing cDNA for purified RNA 10pg derived from HeLa cells using non-amplified reverse transcriptase or this product, and amplifying ACTB by real-time PCR.
Details
Storage condition
Store at -20℃
Components
The kits include the following reagents that can be used for 96 reactions (RMQ-101) or 24 reactions(RMQ-101T). All reagents should be stored at -20℃.
| QuantAccuracy™, RT-RamDA™ cDNA Synthesis Kit | RMQ-101 | RMQ-101T |
|---|---|---|
| Size | 96 Reactions | 24 Reactions |
| Lysis Buffer | 480 μL | 120 μL |
| Lysis Enhancer | 108 μL | 27 μL |
| RNase Inhibitor | 22 μL | 6 μL |
| RT-RamDA™ Buffer | 240 μL | 60 μL |
| gDNA Remover | 54 μL | 14 μL |
| RT-RamDA™ Enzyme Mix | 60 μL | 15 μL |
| RT-RamDA™ Primer Mix | 60 μL | 15 μL |
| Nuclease-free water | 960 μL | 240 μL |
* Do not store mixed solution.
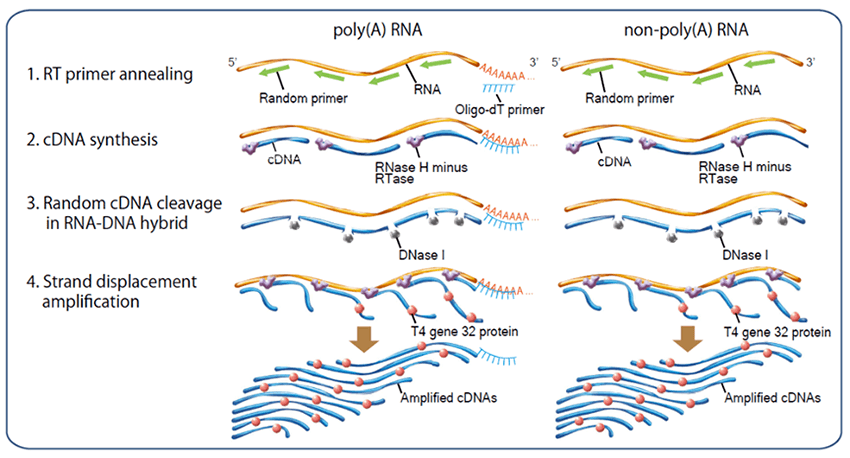
Principle of rt-ramda™ method
RT-RamDA™ is an abbreviation for "Reverse Transcription with Random Displacement Amplification" developed by the Bioinformatics Research and Development Team, RIKEN Center for Biosystems Science and Technology.It is a new cDNA amplification method that applies the strand substitution activity of reverse transcriptase, and it is possible to prepare cDNA not only from poly (A) RNA but also from non-poly (A) -derived RNA for using raondom primer.
In addition, because cDNA is amplified at the same time as it is synthesized, RT-RamDA™ method does not require amplification adaptors and PCR amplification step, and it reduces the biases caused by PCR amplification.
*Hayashi .T et al. Single-cell full-length total RNA sequencing uncovers dynamics of recursive splicing and enhancer RNAs. Nature Communications. 9:619(2018)
Application Data
Example 1.Comparison of the amount of cDNA between the QuantAccuracy™ and conventional Reverse Transcription Kit
cDNA synthesis was performed using 1 ng of total RNA extracted from HeLa S3 cells to compare the QuantAccuracy™, RT-RamDA™ cDNA Synthesis Kit (Code No. RMQ-101) with a conventional reverse transcription kit from Company A.
Real-time PCR analysis was then performed on two types of long non-coding RNA genes (NEAT1 and MALAT1) and eight types of housekeeping genes.
As a result, a correlation of Ct value was observed between this product and the reverse transcription reagent without amplification ability, and ΔCt> 4.5 for all genes, and it was confirmed that this product has an amplification factor of 24.5≒20 times or more.
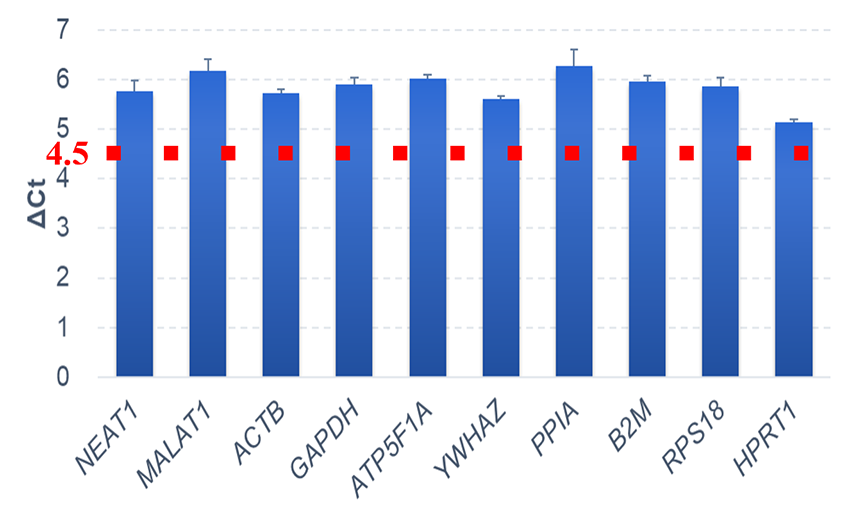
ΔCt values between QuantAccuracy™ and a conventional Reverse Transcription Kit
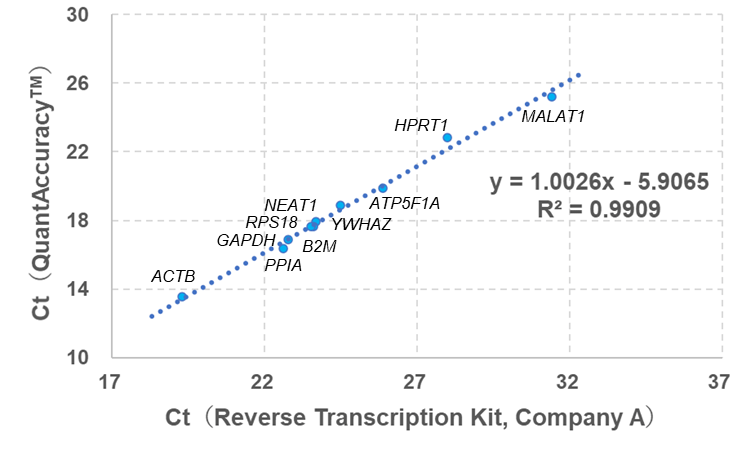
Correlation between QuantAccuracy™ (y-axis) and a conventional Reverse Transcription Kit (x-axis, Company A).
Example 2.Analysis using a human FFPE tissue
<Method>
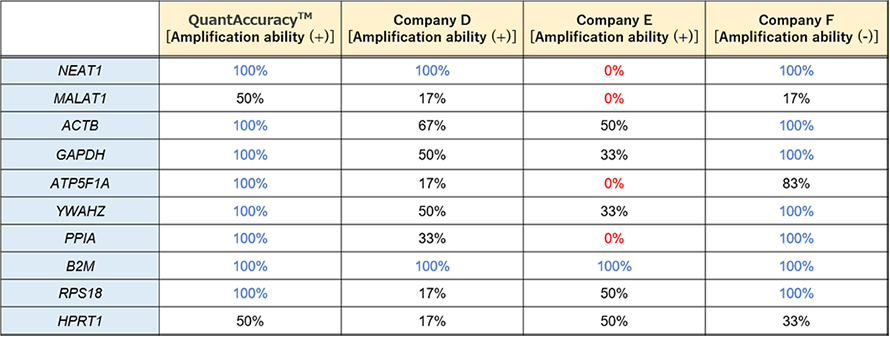
cDNA synthesis was performed using 100 pg or 1 ng RNA extracted from human FFPE tissue to compare the QuantAccuracy™, RT-RamDA™ cDNA Synthesis Kit (Code No.RMQ-101) with other cDNA amplification kits from competitors B and C, and a conventional reverse transcription kit from competitor D. As in Example 1, real-time PCR analysis was performed on two types of long non-coding RNA genes and seven types of housekeeping genes using THUNDERBIRD™ SYBR® qPCR Mix (Code No. QPS-201) with each cDNA used as a template.
The amount of cDNA in the QuantAccuracy™, RT-RamDA™ cDNA Synthesis Kit was calculated from the Ct value, and the value obtained by dividing by the amount of cDNA in the conventional reverse transcription reaction was calculated as the amplification rate.
<Results>
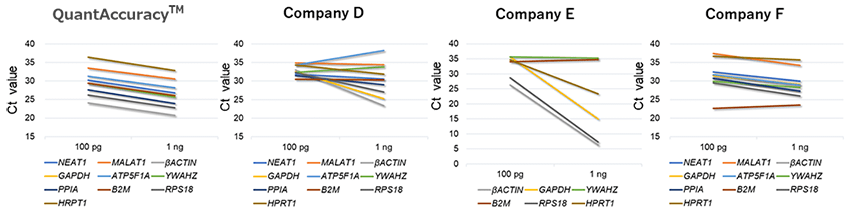
The detection rates of each gene from 100 pg RNA were compared. As a result, cDNA prepared using this product showed the highest detection rate for any of the genes.cDNA was prepared using different RNA amounts (100 pg, 1 ng) as a template, amplified by real-time PCR, and the Ct values were compared.As shown in the figure below, the Ct value when using 100 pg RNA and the Ct value when using 1 ng RNA are connected by a straight line.
There was almost no difference in amplification rate between each gene only when this product was used.In addition, the average ΔCt value between 100pg and 1ng of this product was about 3.40.In other words, in the analysis using this product, the difference in the amount of RNA 10 times can be evaluated as the difference in the amounts of gene 23.4≒ 10.6 times.For RNA derived from FFPE specimens, this product can be analyzed with little bias in amplification factor.
Example 3.Preparation of cDNA from a single cell
<Method>
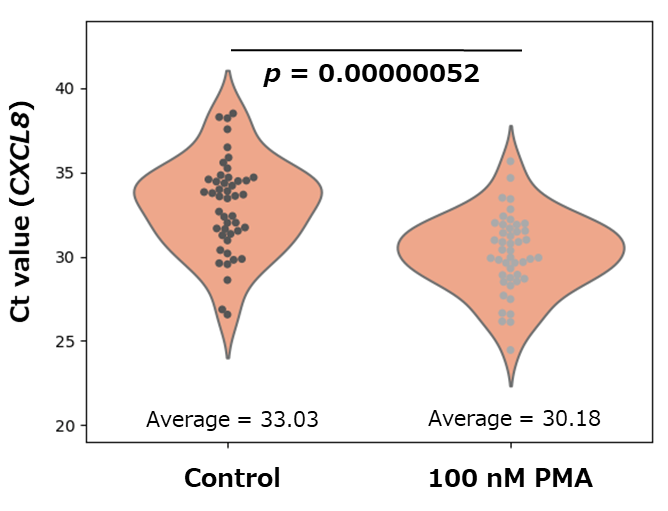
Through Phorbol 12-myristate 13-acetate (PMA) treatment, a single-cell expression analysis experiment of the inflammatory cytokine gene IL-8 was performed. HeLa S3 cells were treated with 100 nM PMA for 24 hours, and then the cells were separated one by one using a cell sorter. cDNA was prepared from one separated cell (n = 48) using this product, and the IL-8 (CXCL8) gene was detected using THUNDERBIRD™ SYBR® qPCR Mix (Code No. QPS-201).
<Results>
It was found that the Ct value was reduced by about 3 by PMA treatment and the expression level of IL-8 was significantly increased (p value was calculated from Student's t-test).By using this product, it was possible to analyze gene expression even in a single cell level.
Example 4.Preparation of cDNA from trace RNA
<Method>
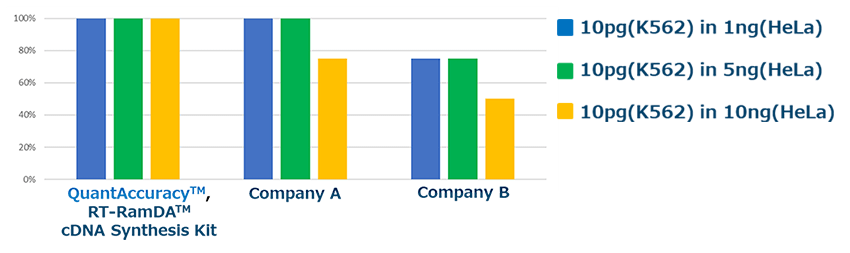
Genes that are thought to be the causative genes of chronic leukemia were investigated.10 pg of K562 cell-derived RNA (BCR-ABL (+)) was spiked into 1 ng, 5 ng, and 10 ng HeLa cell-derived RNA (BCR-ABL (-)). Then, cDNA was prepared using each reverse transcription reagent, and the BCR-ABL gene was detected by real-time PCR using THUNDERBIRD™ SYBR® qPCR Mix (Code No. QPS-201) (n = 4).
<Results>
As the amount of RNA derived from HeLa cells increased, the efficiency of cDNA synthesis from trace of RNA decreased with the reverse transcription reagent without amplification ability, and detection omission occurred. Our products were able to efficiently prepare cDNA and all were detectable.
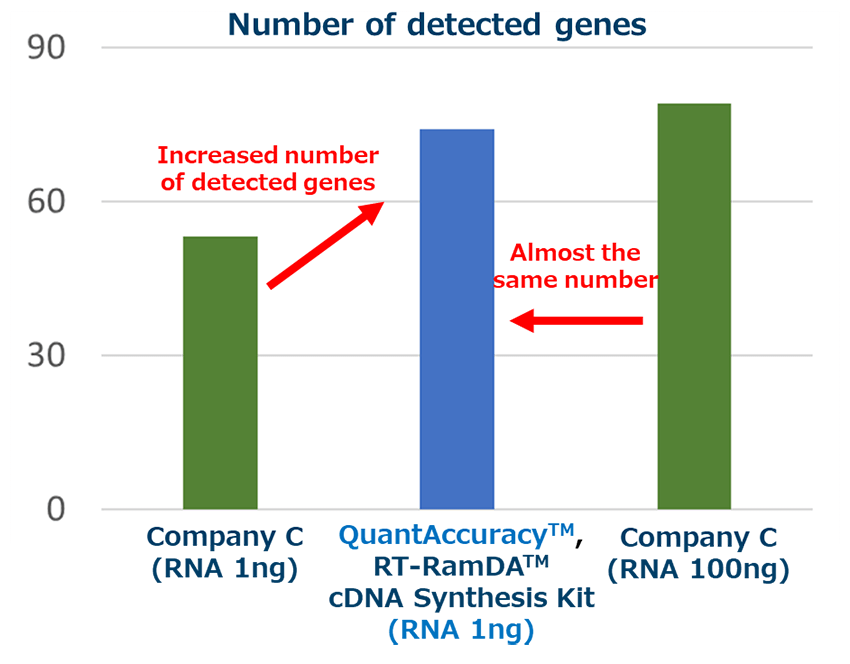
Example 5.Input amount and number of detected genes
<Method>
cDNA was prepared from iPS cell-derived RNA (1 ng, 100 ng) using each reverse transcription reagent. Then, it was added to TaqMan Array Mouse Stem Cell Pluripotency, and 96 kinds of genes were detected by THUNDERBIRD™ Probe qPCR Mix (Code No. QPS-101).
<Results>
In the input amount of RNA1ng, this product was able to detect more genes than the reagent of Company C. In addition, almost the same number of genes were detected as when cDNA was prepared from 100 times the amount of RNA (100 ng) using the company C reverse transcription reagent.