- HOME
- Single Cell/Ultra-low amount RNA GenNextTM RamDA-seqTM Single Cell Kit
-
Single Cell/Ultra-low amount RNA
GenNextTM RamDA-seqTM Single Cell Kit
Code No. RMD-101T, RMD-101
DESCRIPTION
-
-
GenNext™ RamDA-seq™ Single Cell Kit is a kit using RT-RamDA™ (Reverse Transcription with Random Displacement Amplification) method for preparing cDNA for NGS analysis from single-cell and Ultra-low amount RNA.
RT-RamDA™ is a novel cDNA amplification method utilizing the strand displacement activity of reverse-transcriptase. GenNext RamDA-seq™ Single Cell Kit can detect not only poly(A) RNA but also non-poly(A) with high sensitively by using random primer or NSR primer*, allowing the detection of many genes compared with the conventional method.
Unlike conventional method using Oligo-dT primer, cDNA for full-length total RNA-seq can be synthesized by RT-RamDA™ method.
In addition, because cDNA is amplified at the same time as it is synthesized, RamDA-seq™ method does not require amplification adaptors and PCR amplification step, and it reduces the biases caused by PCR amplification.
-

* NSR primer
NSR primer is abbreviation for Not so random primer. NSR primer is designed random primer to avoid synthesizing cDNA from the rRNAs by removing 6-mers that exactly match the rRNA sequences from N6 random primers.
In order to perform more efficient NGS analysis, it is necessary to reduce the rRNA contamination rate.As a countermeasure, we also offer Not so random primers for humans and mice sold separately.
* RamDA-seq™ and RT-RamDA™ are a trademark of RIKEN, Institute of Physical and Chemical Research
RT-RamDA™ cDNA Synthesis kit for qPCR
- Here -
Features
- - cDNA can be prepared from a single cell or small amount of input RNA
1-100 cells or 10 pg-1 ng total RNA - - Full length cDNA can be analyzed
cDNA that covers the entire length of the target RNA of 10 kb or more can be prepared. - - Various RNAs can be detected, and the number of genes detect able is higher than that with conventional technology.
- · Identification of isoforms and alternative splicing
- · Detection of poly(A) RNA and non poly(A) RNA (histone RNA and lncRNA)
- · Detection of nuclear RNA (pre mRNA and lncRNA)
Details
STORAGE CONDUTION
Store at-20 ℃
COMPONENTS
The kits include the following reagents that can be used for 96 (RMD-101) and 24 (RMD-101T) reactions. All reagents should be stored at −20℃.
| GenNext™ RamDA-seq™ Single Cell Kits |
RMD-101 | RMD-101T |
|---|---|---|
| Size | 96 Rxns | 24 Rxns |
| Lysis Buffer | 480 μL | 120 μL |
| Lysis Enhancer | 108 μL | 27 μL |
| RNase Inhibitor | 22 μL | 6 μL |
| Nuclease free water | 960 μL | 240 μL |
| RT-RamDA™ Buffer | 240 μL | 60 μL |
| RT-RamDA™ Enzyme Mix | 54 μL | 14 μL |
| RT-RamDA™ Primer Mix | 54 μL | 14 μL |
| gDNA Remover | 54 μL | 14 μL |
| 2nd strand synthesis Buffer | 330 μL | 83 μL |
| 2nd strand synthesis Enzyme | 55 μL | 14 μL |
| 2nd strand synthesis Primer Mix | 275 μL | 69 μL |
*Do not store mixed solution.
| NSR Primer Set for human | NSR-101 |
|---|---|
| 1st NSR Primer Mix for human | 54 μL |
| 2nd NSR Primer Mix for human | 275 μL |
| NSR Primer Set for mouse | NSR-102 |
|---|---|
| 1st NSR Primer Mix for mouse | 54 μL |
| 2nd NSR Primer Mix for mouse | 275 μL |
Required materials not included
- · Thermocycler
- · Library preparation reagents
GenNext RamDA-seq™ Single Cell Kits are specifically for use with the Illumina Nextera™ XT DNA Sample Kit. - · Nextera™ XT DNA Sample Kit (24 Samples), Cat. no. FC-131-1024
- · Nextera™ XT DNA Sample Kit (96 Samples), Cat. no. FC-131-1096
- · SPRI (Solid Phase Reversible Immobilization) paramagnetic beads
- · Agencourt™ AMPure™ XP Beads (Beckman Coulter, Cat. no. A63880 or A63881)
- · TE Buffer pH 8.0 (10 mM Tris-HCl,,1mM EDTA)
- · Magnetic rack/stand for magnetic bead separation
- · 80% Ethanol (freshly prepared)
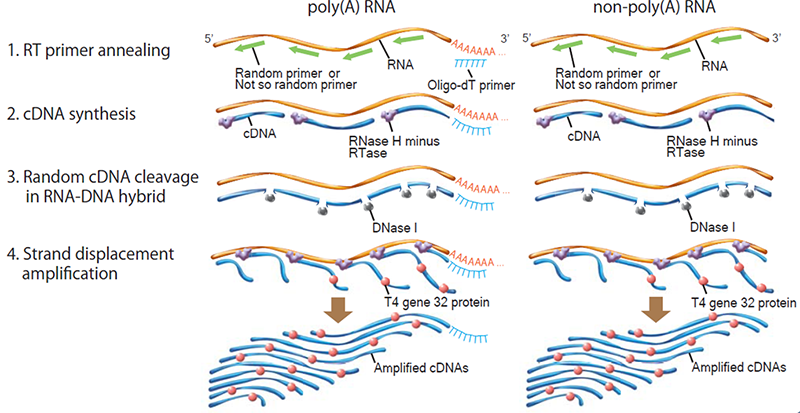
Principle of RT-RamDA™ method
RT-RamDA™ is an abbreviation for "Reverse Transcription with Random Displacement Amplification" developed by the Bioinformatics Research and Development Team, RIKEN Center for Biosystems Science and Technology.It is a new cDNA amplification method that applies the strand substitution activity of reverse transcriptase, and it is possible to prepare cDNA not only from poly (A) RNA but also from non-poly (A) -derived RNA for using raondom primer.
Unlike conventional method using Oligo-dT primer, cDNA for full-length total RNA-seq can be synthesized by RT-RamDA™ method.
In addition, because cDNA is amplified at the same time as it is synthesized, RT-RamDA™ method does not require amplification adaptors and PCR amplification step, and it reduces the biases caused by PCR amplification.
* Hayashi .T et al. Single-cell full-length total RNA sequencing uncovers dynamics of recursive splicing and enhancer RNAs. Nature Communications. 9:619(2018)
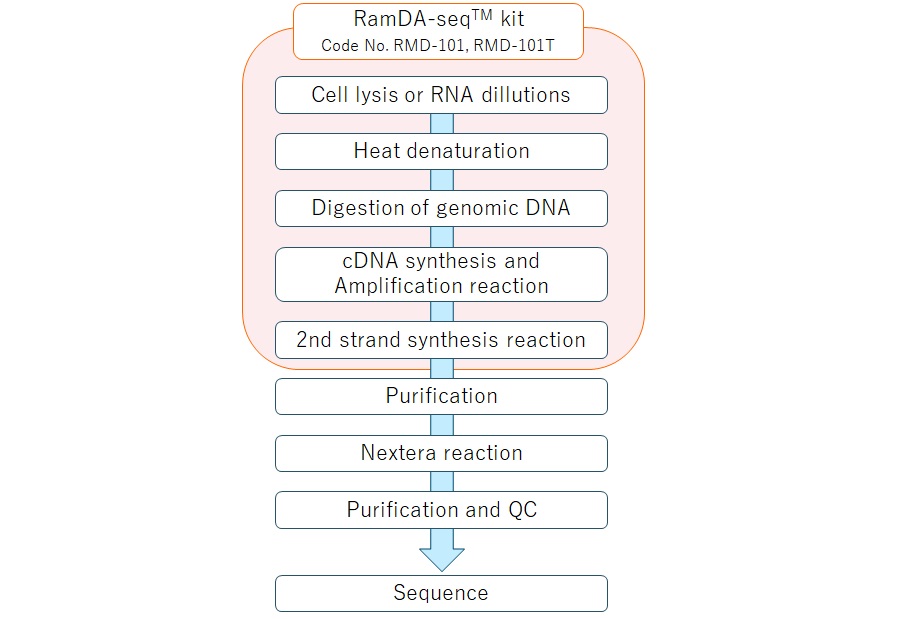
Workflow
Application Data
Example 1.Comparison of the number of detected transcripts and the alignment ratio within each region
Using next generation sequencing data, cDNA and double-stranded DNA were prepared using either the GenNext™ RamDA-seq™ Single Cell Kit (Code No. RMD-101) or a Company A kit, with a sample input of 10 pg of total RNA extracted from mouse ES cells. The GenNext™ RamDA-seq™ Single Cell Kit uses the NSR Primer Set for mouse (code No.NSR-102). PCR for cDNA amplification, which is not required with the GenNext™ RamDA-seq™ Single Cell Kit, was performed for 18 cycles with the Company A kit. The libraries were then prepared with the Nextera™ XT DNA Library Preparation Kit and sequenced with the Illumina MiSeq instrument. Thus, the GenNext™ RamDA-seq™ Single Cell Kit was shown to detect approximately 5,000 more genes than the Company A kit.

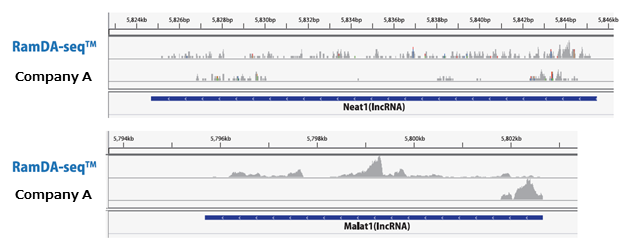
Example 2.Comparison of the full-length coverage of long RNA molecules
Analysis was performed on the sequence data obtained with the method described for Example 1. The GenNext™ RamDA-seq™ Single Cell Kit can detect lncRNAs such as Neat1 and Malat1, which were difficult to capture with the Company A kit over their entire length.
Example 3.Comparison of the rate of rRNA contamination using random Primers or NSR Primers
Using the RT-RamDA™ Primer Mix(random Primers) included with the GenNext™ RamDA-seq™ Single Cell Kit [Code No. RMD-101] or the separately sold NSR Primer Set for mouse [Code No. NSR-102], cDNA and double-stranded DNA were synthesized from total RNA (10pg, 1ng) extracted from NIH3T3 cells. libraries were prepared using the Nextera™ XT DNA Library Preparation Kit and Sequencing was performed on an Illumina™ MiSeq™ instrument.
For both 10pg and 1ng of Total RNA, NSR Primers resulted in the lower rate of rRNA contamination compared to random primers.
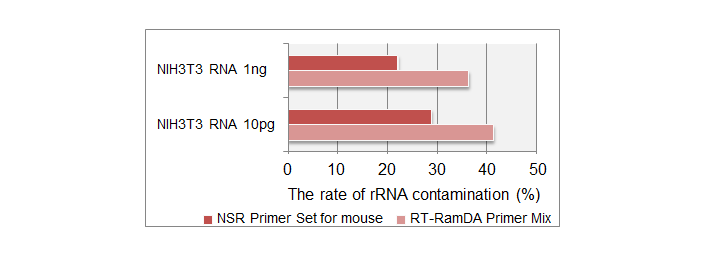
Using NSR primers can reduce the rate of rRNA contamination more than random primers.
For human-derived samples, it is recommended to use the NSR Primer Set for human [Code No. NSR-101], and for mouse-derived samples, the NSR Primer Set for mouse [Code No. NSR-102]
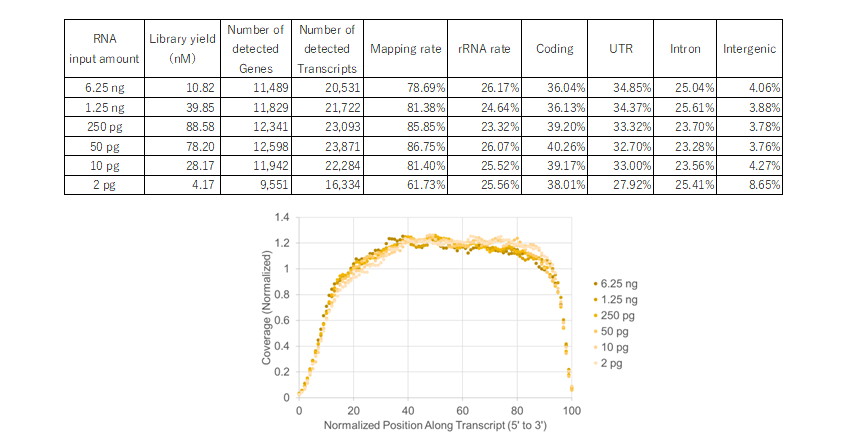
Example 4.RNA-seq analysis from various RNA input amounts (2 pg to 6.25 ng)
Using RNA extracted from HeLa cells, libraries were prepared with input amounts ranging from 2 pg to 6.25 ng. For cDNA synthesis, the NSR Primer Set for human [Code No. NSR-101] was used, and all library enrichments were performed with 15 cycles.Sequencing was performed on an Illumina™ MiSeq™ instrument, and NGS analysis was performed using basespace (Illumina) RNA-Seq Alignment (ver. 2.0.2) or RamDAQ (ver.1.9).
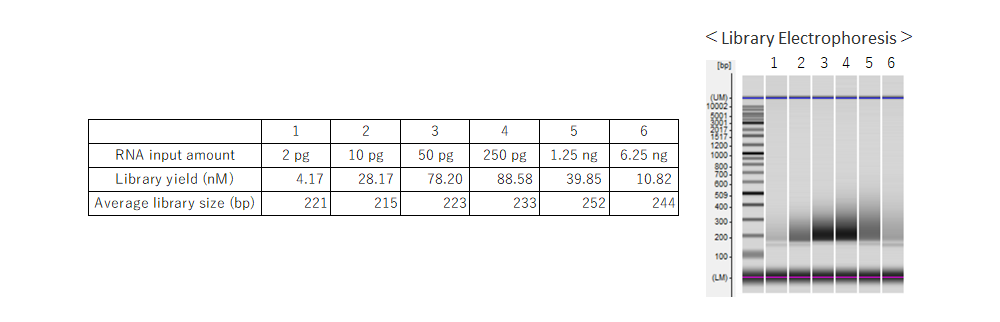
The number of detected transcripts and genes was low at 2 pg because the input amount was low to begin with. The number of detected transcripts and genes was high at input amounts other than 2 pg, and the coverage was unbiased at all input amounts.
Publications Using GenNext™ RamDA-seq™ Single Cell Kit
Yoshimatsu, S. et al., Non-viral Induction of Transgene-free iPSCs from Somatic Fibroblasts of Multiple Mammalian Species. Stem Cell Reports, Volume 16:754-770(2021)
Hasegawa, N. et al., Highly sensitive fusion detection using plasma cell-free RNA in non-small-cell lung cancers. Cancer Sci. 112(10):4393-4403(2021)
Oka, Y. et al., A novel sorting signal for RNA packaging into small extracellular vesicles. Scientific Reports 13, Article number: 17436 (2023)
Usuda, M. et al., Inhibitory Effect of Adsorption of Streptococcus mutans onto Scallop-Derived Hydroxyapatite. Int. J. Mol. Sci. 24,11371 (2023)
