- HOME
- Single Cell/Ultra-low amount RNA GenNextTM Shin-RamDA-seqTM Single Cell Stranded Kit
-
Single Cell/Ultra-low amount RNA
GenNextTM Shin-RamDA-seqTM Single Cell Stranded Kit
Code No. RML-101T, RML-101
*This product is not available in the U.S. and Europe.
DESCRIPTION
-
-
GenNext™ Shin-RamDA-seq™ Single Cell Stranded Kit is an improved version of the GenNext™ RamDA-seq™ Sigle Cell Kit, and is a kit for preparing libraries for next-generation sequencing (NGS) from single cell or trace RNA. Obtained libraries could be applied to subsequent strand specific RNA-seq analysis. By using this kit, you can prepare NGS libraries that cover the full length of RNA and also perform strand analysis.
This is a kit using RT-RamDA™ (Reverse Transcription with Random Displacement Amplification) method developed by RIKEN. RT-RamDA™ is a novel cDNA amplification method utilizing the strand displacement activity of reverse-transcriptase. This kit can detect not only poly(A) RNA but also non-poly(A) with high sensitively by using random primer or NSR primer*.
Additionally, Shin-RamDA-seq™ incorporates further reduction of rRNA and preservation of strand information, enabling more accurate RNA-Seq analysis.
* NSR primer is abbreviation for Not so random primer.
* Shin-RamDA-seq™ is an abbreviation for "Stranded High-Sensitivity Random Displacement Amplification sequencing".
* Shin-RamDA-seq™, RamDA-seq™ and RT-RamDA™ are a trademark of RIKEN, Institute of Physical and Chemical Research
-
Features
- - Compatible with single cell or trace RNA
cDNA can be prepared from 1-100 cells or 10pg-1ng total RNA. - - Capable of full length RNA analysis
Long-chain RNA and splicing variants can be accurately analyzed. - - Capable of analyzing total RNA
This kit can be used for non-poly(A)RNA (lncRNA, pre-mRNA, circRNA, etc.) and degraded RNA(ex. FFPE-derived RNA). - - Accurate analysis is made possible by preserving strand information
Antisense and overlapping genes can be found and clearly distinguished. - - Support for library preparation
Library preparation reagents support all steps from cDNA synthesis to library construction. (Magnetic beads for purification and index kit are required separately.)
Details
STORAGE CONDUTION
Store at -20℃
COMPONENTS
The kits include the following reagents that can be used for 96 (RML-101) and 24 (RML-101T) reactions. All reagents should be stored at −20℃.
| GenNext™ Shin-RamDA-seq™ Single Cell Stranded Kit |
RML-101 | RML-101T |
|---|---|---|
| Size | 96 Rxns | 24 Rxns |
| Lysis Buffer | 240 μL | 60 μL |
| Lysis Enhancer | 240 μL | 60 μL |
| RNase Inhibitor | 22 μL | 6 μL |
| Nuclease free water | 960 μL | 240 μL |
| RT-RamDA™ Buffer | 240 μL | 60 μL |
| gDNA Remover | 54 μL | 14 μL |
| rRNA Remover | 30 μL | 8 μL |
| RT-RamDA™ Enzyme Mix | 54 μL | 14 μL |
| RT-RamDA™ Primer Mix | 54 μL | 14 μL |
| 2nd strand synthesis Buffer | 330 μL | 83 μL |
| 2nd strand synthesis Enzyme | 55 μL | 14 μL |
| 2nd strand synthesis Primer Mix | 275 μL | 69 μL |
| Fragmentase | 120 μL | 30 μL |
| End Repair and A-tailing Buffer | 96 μL | 24 μL |
| End Repair and A-tailing Enzyme | 24 μL | 6 μL |
| Ligation Solution | 480 μL | 120 μL |
| Library Amplification Master Mix | 300 μL | 75 μL |
| Library Amplification Primer Mix | 120 μL | 30 μL |
*Do not store mixed solution.
*RamDA Cell Lysis Kit [Code No. RMD-301] is not compatible with this product.
Option:The following products are useful for depletion of human or mouse rRNA. (NSR Primer sold separately.)
| NSR Primer Set for human | NSR-101 |
|---|---|
| 1st NSR Primer Mix for human | 54 μL |
| 2nd NSR Primer Mix for human | 275 μL |
| NSR Primer Set for mouse | NSR-102 |
|---|---|
| 1st NSR Primer Mix for mouse | 54 μL |
| 2nd NSR Primer Mix for mouse | 275 μL |
Required materials not included
- · Thermocycler
- · Index kit
(Recommend) IDT for Illumina – TruSeq DNA UD Indexes v2 (96 Indexes,96 Samples), (Illumina, Inc. Cat. no. 20040870) - · SPRI (Solid Phase Reversible Immobilization) paramagnetic beads
Agencourt™ AMPure™ XP Beads (Beckman Coulter, Cat. no. A63880 or A63881) - · TE Buffer pH 8.0 (10 mM Tris-HCl,1mM EDTA)
- · 10 mM Tris-HCl pH 8.0
- · Magnetic rack/stand for magnetic bead separation
- · 80% Ethanol (freshly prepared)
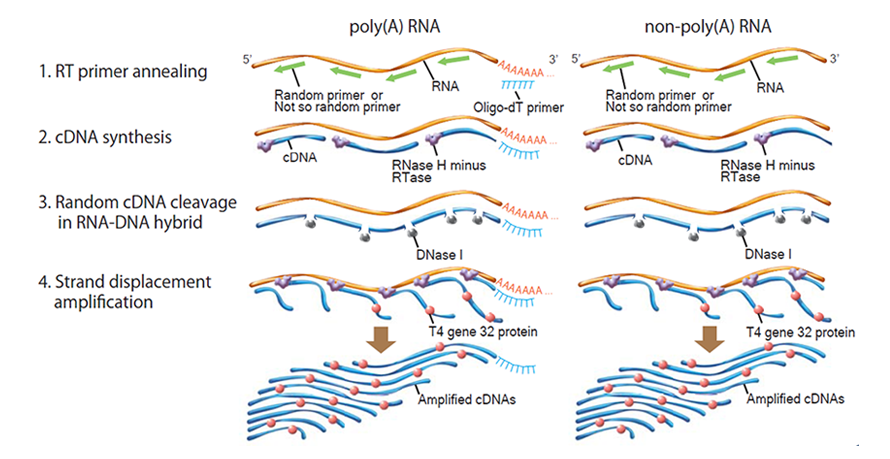
Principle of RT-RamDA™ method
RT-RamDA™ is an abbreviation for "Reverse Transcription with Random Displacement Amplification" developed by the Bioinformatics Research and Development Team, RIKEN Center for Biosystems Science and Technology.It is a new cDNA amplification method that applies the strand substitution activity of reverse transcriptase, and it is possible to prepare cDNA not only from poly (A) RNA but also from non-poly (A) -derived RNA for using raondom primer.
Unlike conventional method using Oligo-dT primer, cDNA for full-length total RNA-seq can be synthesized by RT-RamDA™ method.
In addition, because cDNA is amplified at the same time as it is synthesized, RT-RamDA™ method does not require amplification adaptors and PCR amplification step, and it reduces the biases caused by PCR amplification.
* Hayashi .T et al. Single-cell full-length total RNA sequencing uncovers dynamics of recursive splicing and enhancer RNAs. Nature Communications. 9:619(2018)
Application Data
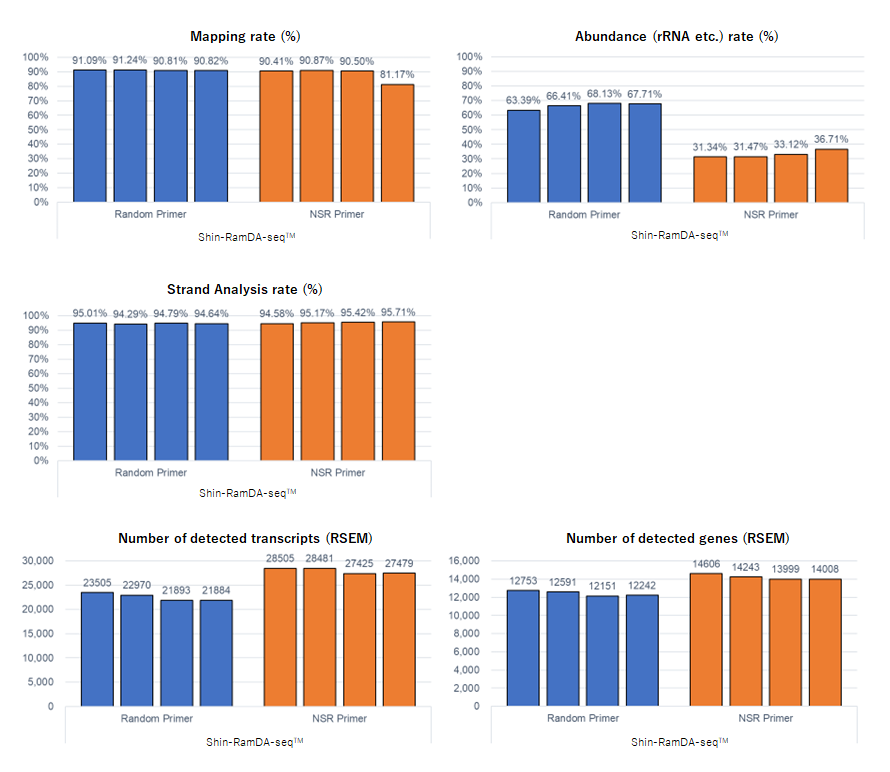
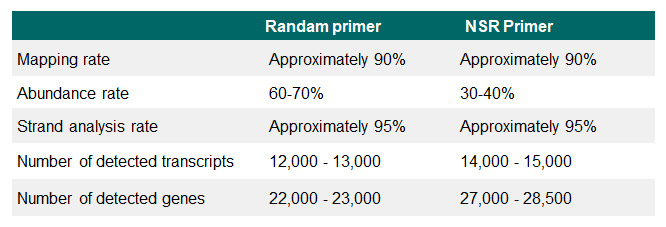
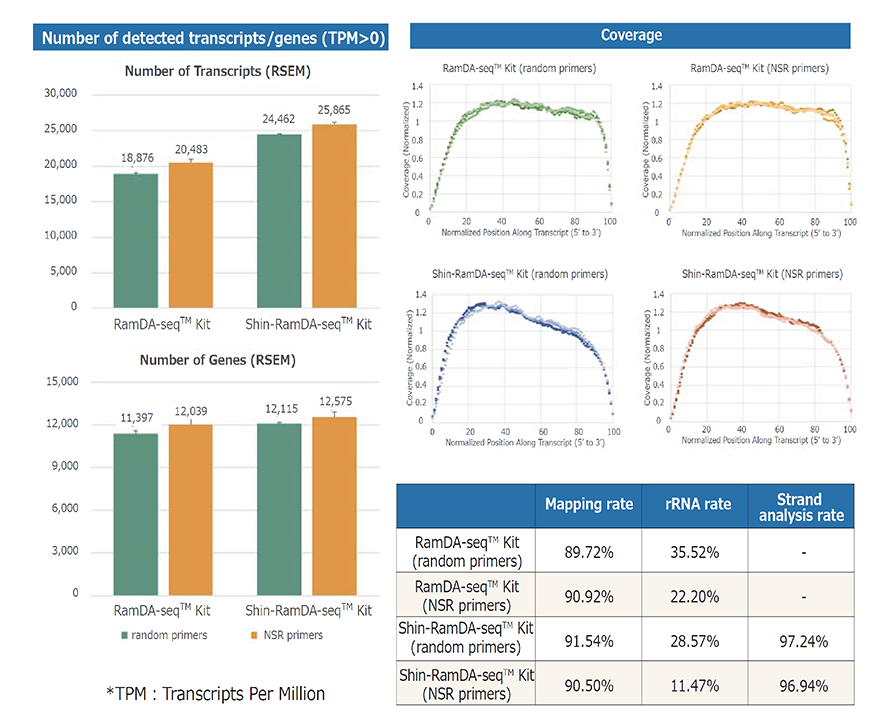
Example 1.Analysis of NIH3T3 cells (Shin-RamDA-seq™ Kit vs. RamDA-seq™ Kit)
NGS libraries were prepared from single cell quantity extracts from mouse-derived NIH3T3 cell and subjected to RNA-Seq analysis. The number of detected transcripts/genes, average coverage, mapping rate, rRNA rate, and strand analysis rate were analyzed. Illuminaʼs MiSeq™ was used for RNA-Seq.
The Shin-RamDA-seq™ Kit demonstrated a high strand analysis rate and preserved RNA direction information. Optimizing Shin-RamDA-seq™ Kit reagent composition resulted in lower rRNA contamination than that of the RamDA-seq™ Kit, and the number of detected genes was higher than was obtained using a RamDA-seq™ Kit. The use of NSR primers reduced rRNA contamination and increased the number of genes detected.
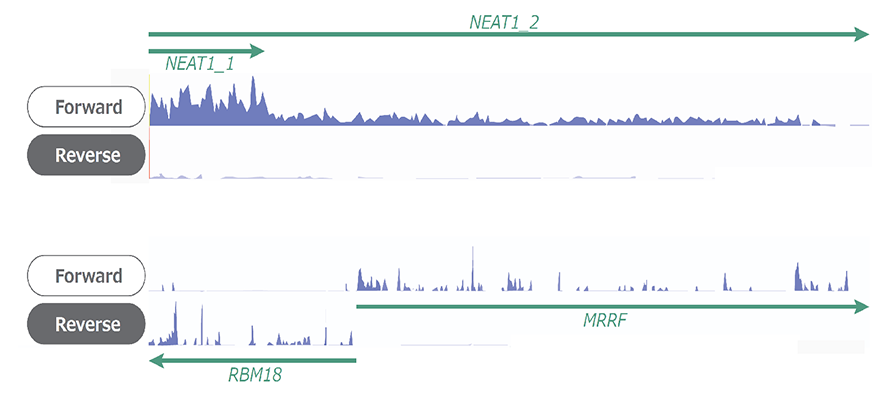
Example 2.Read coverage using Shin-RamDA-seq™ Kit
NGS libraries were prepared from single cell quantity extracts from the K562 human leukemia cell line, and RNA-seq analysis was performed. Coverage was confirmed using Millefy*. Single-cell analysis using a Shin-RamDA-seq™ Kit confirmed full-length coverage of NEAT1_2 with 20 kb lengths and allowed isoform analysis of NEAT1_1 and NEAT1_2. Preservation of strand information enabled precise analysis of MRRF and RBM18, which are known to be transcribed in opposite direction.
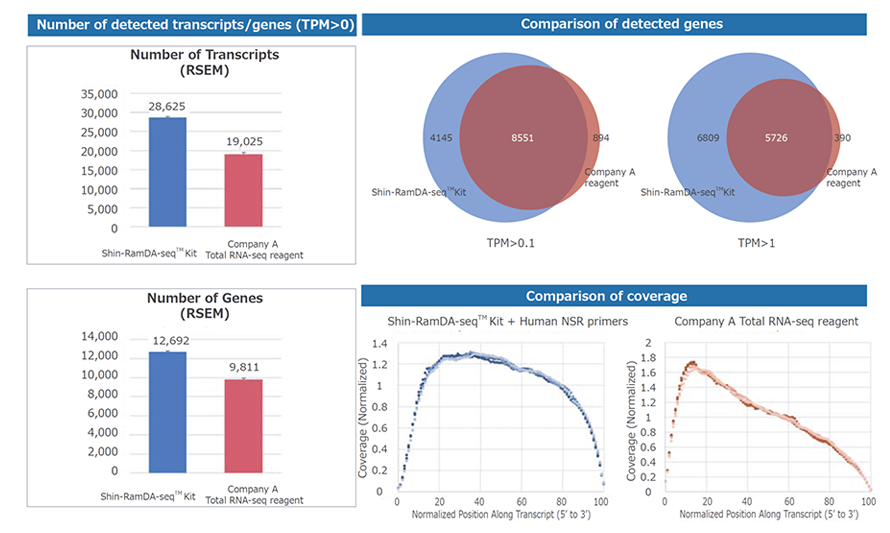
Example 3.Analysis of K562 cells (Comparison of Shin-RamDA-seq™ and another company's reagent)
Using a Shin-RamDA-seq™ Kit or another company's Total RNA-seq reagent, NGS libraries were prepared from single cell quantities of human K562 cell RNA extractions. NGS analysis was performed using Illumina's MiSeq™.
Numbers of detected transcripts/genes and coverage were compared with another company's reagent. The Shin-RamDA-seq™ Kit showed a higher number of detected transcripts/genes, and the Shin-RamDA-seq™ Kit provided more unbiased results in terms of coverage.
Example 4.Analysis of FFPE sample
From 1ng of FFPE-derived RNA, library preparation was performed using the GenNext™ Shin-RamDA-seq™ Single Cell Stranded Kit [Code No. RML-101], and RNA-Seq was performed. Illumina MiSeq™ was used for RNA-Seq. The preparation and analysis conditions for FFPE and RNA-Seq are as follows.
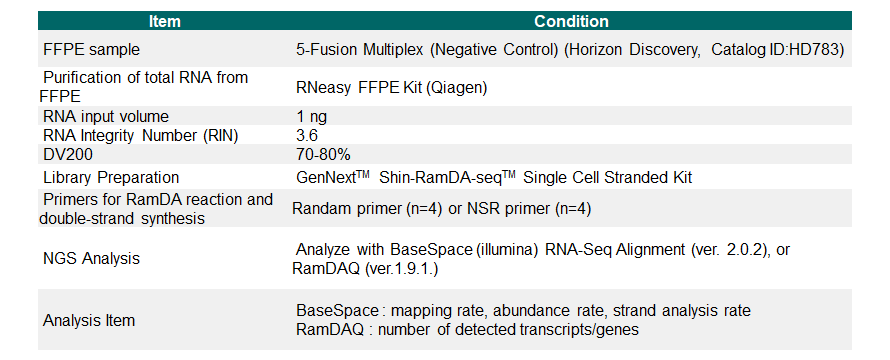
Each analysis resulted in the detection of over 10,000 genes by using the GenNext™ Shin-RamDA-seq™ Single Cell Stranded Kit. Furthermore, by using the NSR primer [Code No. NSR-101], which is a random primer designed to exclude sequences that perfectly match the 18S and 28S RNA genes, the detection of rRNA was suppressed, allowing for more accurate analysis.