- HOME
- GenNextTM NGS Library Prep Kit -KODEasyTM-
-
GenNextTM NGS Library Prep Kit -KODEasyTM-
Code No.LPK-201T, LPK-201, LPK-201L
DESCRIPTION
-
-
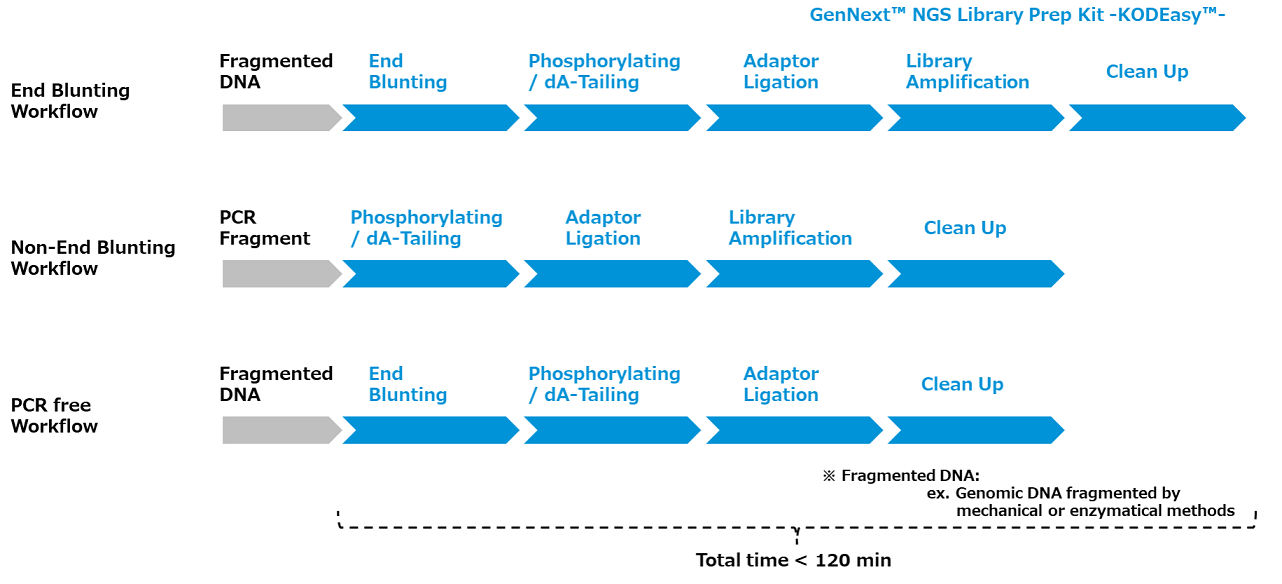
GenNext™ NGS Library Prep Kit -KODEasy™- comprises the enzymes and buffers for preparing libraries for Illumina® sequencing from fragmented double-stranded DNA and PCR products. This ligation-based library preparation system offers a simplified workflow by enabling direct amplification of NGS libraries without requiring a purification step after adapter ligation. Terminal repair and 3′ end adenylation of the fragmented DNA can be conducted in the End Blunting and Phosphorylating & dA-Tailing step. Platform-specific adapters are then ligated to both ends of the DNA fragments. If required, PCR-free workflow is also available.
-
Features
- Improved usability
Reagent volumes ≥ 3 μL simplify pipetting, facilitate automation and enhance reproducibility.
- Low-bias amplification
A proprietary PCR enzyme (UKOD Polymerase) ensure efficient amplification of both AT-rich and GC-rich targets, enabling low-bias library amplification.
- Single-tube reaction
All steps from end-repair to library amplification are performed in a single tube, minimizing handling errors and simplifying the workflow.
- Only one purification step
A singe purification step simplifies the workflow, reduces sample loss, and conserves consumables, making it suitable for high-throughput processing. Sample loss is minimized, and the use of consumables such as tips is significantly reduced.

Details
STORAGE CONDITION
Store at -20°C
COMPONENTS
The kits include the following reagents, which can be used for 8 (LPK-201T), 24 (LPK-201) and 96 (LPK-201L) reactions. All reagents should be stored at -20°C.
| GenNext™ NGS Library Prep Kit -KODEasy™- | LPK-201T | LPK-201 | LPK-201L |
|---|---|---|---|
| Size | 8 Rxns | 24 Rxns | 96 Rxns |
| KOD Blunting Solution (KB solution) |
56 μL | 168 μL | 672 μL |
| Phosphorylating & A-Tailing Solution (PAT Solution) |
24 μL | 72 μL | 288 μL |
| Ligation Solution | 120 μL | 360 μL | 720 μl × 2 |
| Library Amplification Solution | 160 μL | 480 μL | 960 μl × 2 |
| Library Amplification Primer Mix | 40 μL | 120 μL | 480 μL |
*Adapters and magnetic beads for purification are not included in this product.
Workflow
Molecular mechanism of GenNext™ NGS Library Prep Kit -KODEasy™-
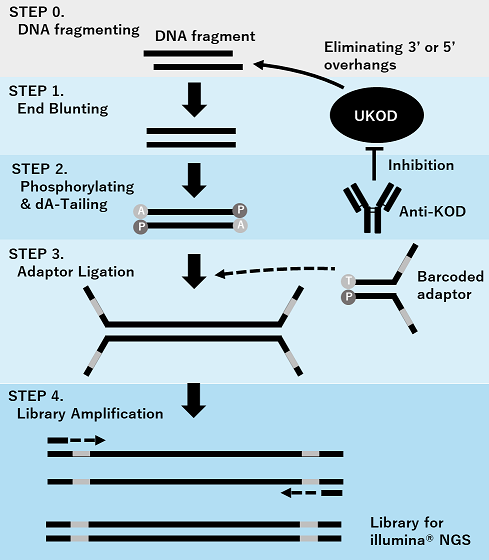
STEP 1. Use a genetically modified KOD DNA polymerase (UKOD), which exhibits high fidelity and amplification efficiency, as the end-blunting enzyme.
STEP 2. Inhibit the activity of the end-blunting enzyme using two types of monoclonal antibodies and simultaneously perform 5′ phosphorylation and dA-tailing.
STEP 3. Enzyme suppression minimizes adapter dimer formation and allows for efficient adapter ligation.
STEP 4. Amplify the library using a highly specific hot-start PCR with UKOD.
Application Data
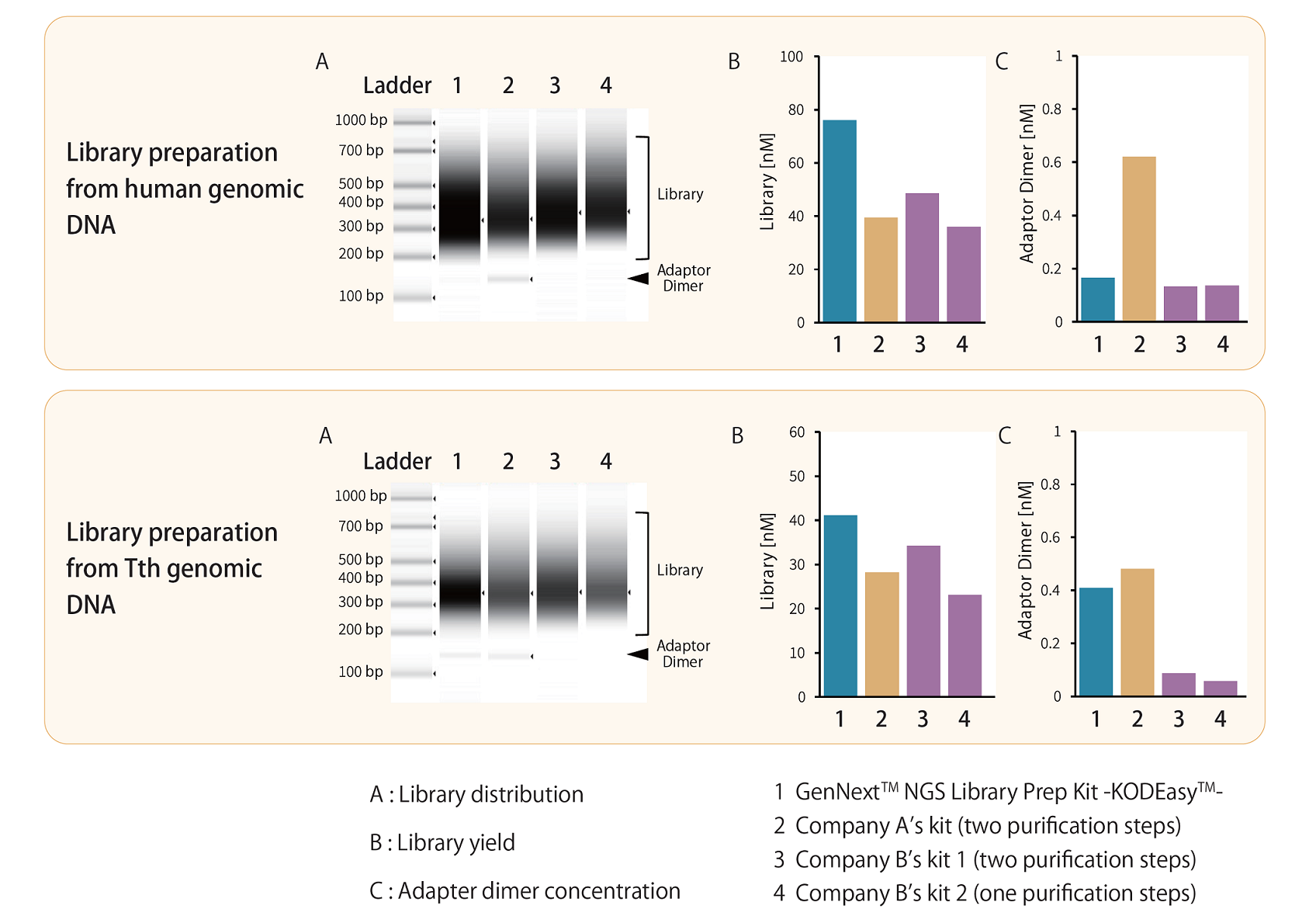
Example 1.Comparison of library distribution and yield
Libraries were prepared from 10 ng of sonicated human genomic DNA (average fragment size: 200 bp) and Thermus thermophilus HB8 (Tth) genomic DNA using the GenNext™ NGS Library Prep Kit -KODEasy™- and competitor kits. Library size distribution, yield, and adapter dimer concentration were compared. PCR was performed for 8 cycles.
The results showed comparable library size distributions equal or higher yields with GenNext™ NGS Library Prep Kit -KODEasy™-, and similar adapter dimer concentrations. All libraries achieved yields exceeding 4 nM, sufficient for Illumina sequencing.
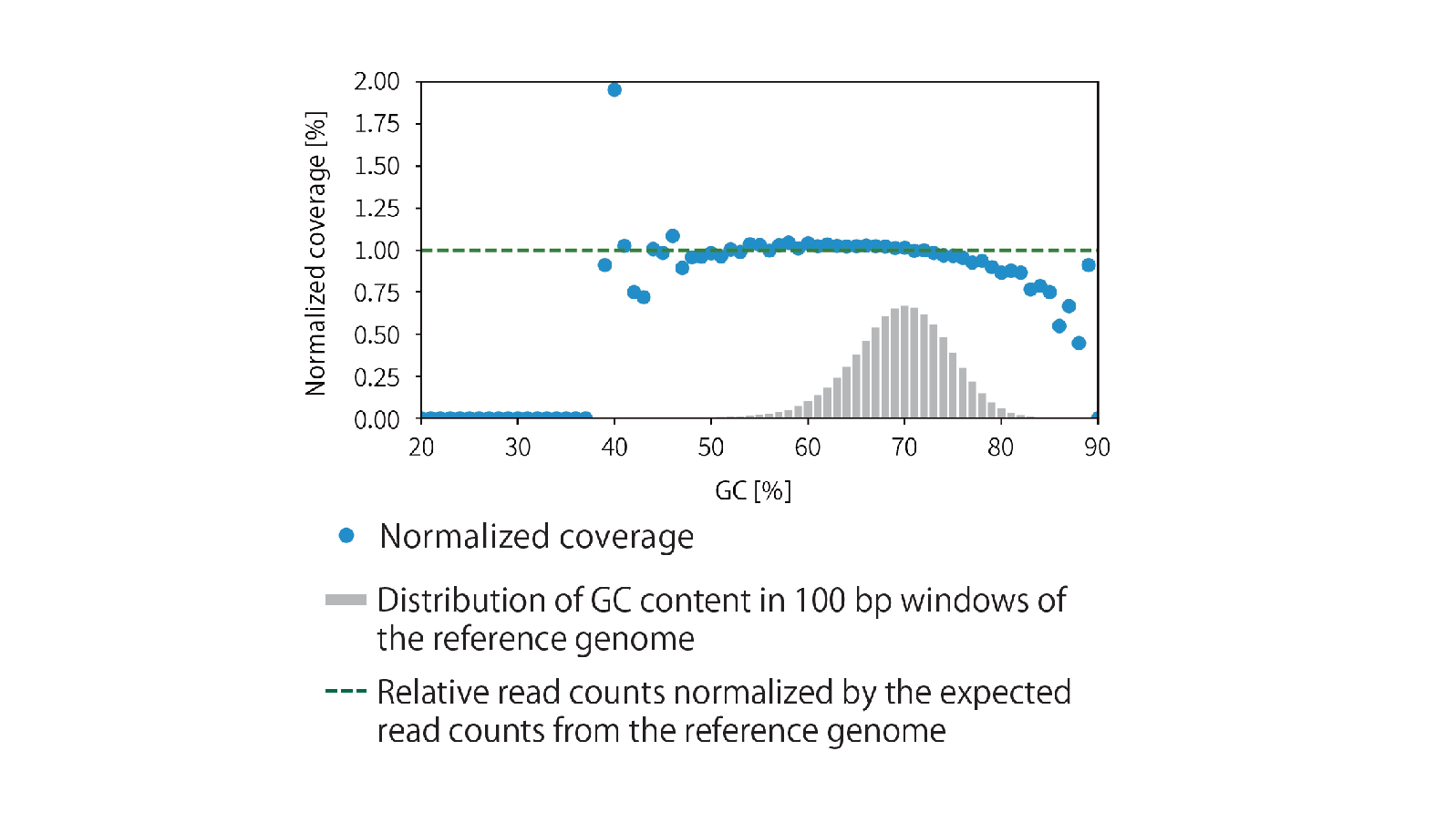
Example 2.Evaluation of uniformity in library coverage
The library was prepared from 10 ng of sonicated Thermus thermophilus HB8 (Tth) genomic DNA (average chain length 200 bp) using the GenNext™ NGS Library Prep Kit -KODEasy™-, and the coverage in GC ratio was confirmed.
The library was amplified for 8 cycles, and sequencing was performed using MiSeq (Illumina). High detection efficiency was maintained even in genomic regions with a GC content exceeding 70%.
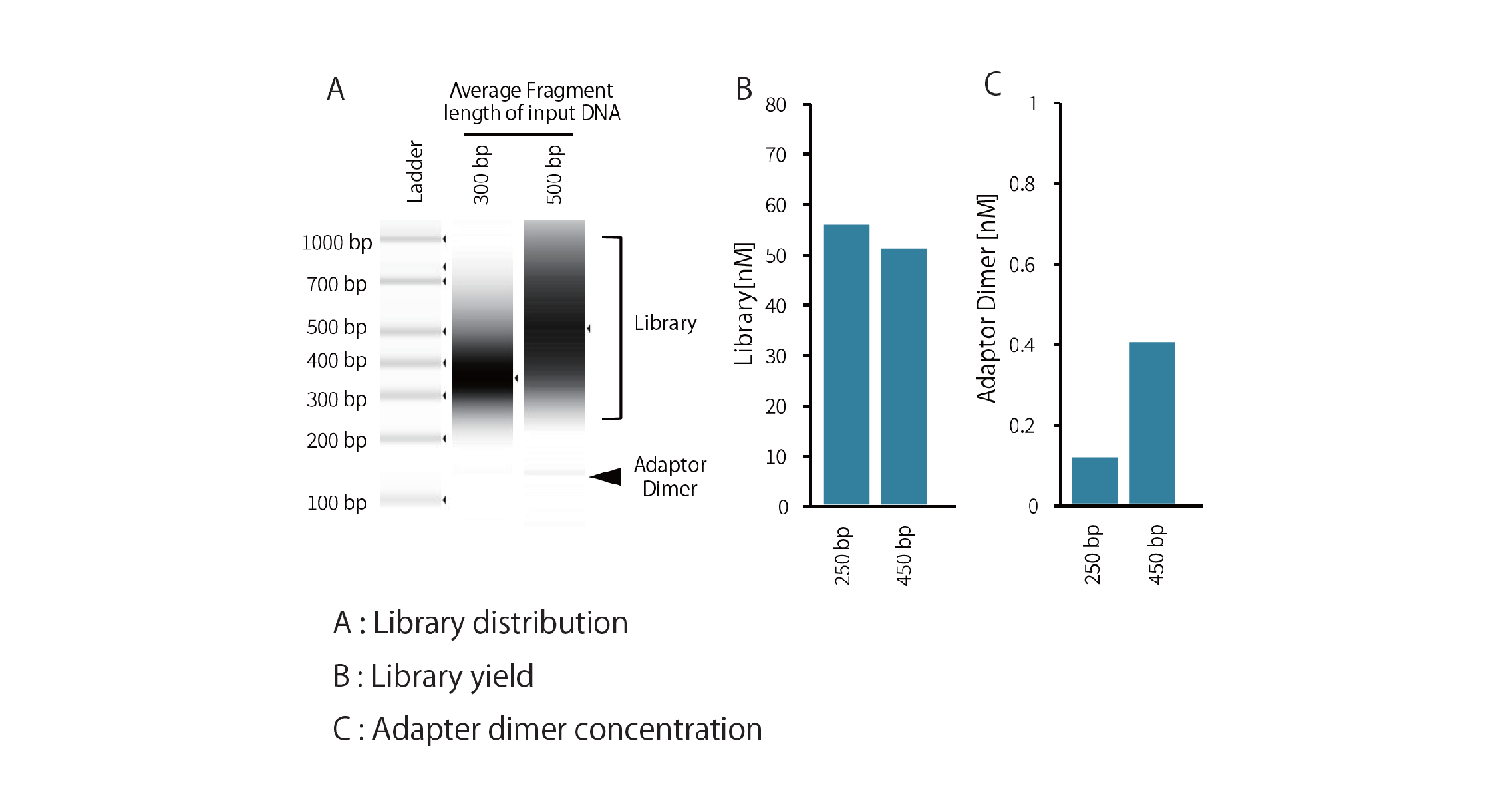
Example 3.Library preparation from input DNA with various average fragment lengths
Libraries were prepared from 10ng of human genomic DNA fragmented by ultrasonication (average chain length 300bp and approximately 500bp) using the GenNext™ NGS Library Prep Kit -KODEasy™-, and library distribution, yield, and adapter dimer concentration were compared. The PCR cycle number was 8 cycles.
Regardless of input DNA fragment length, all preparations yielded ≥4nM, which is sufficient for Illumina sequencing.
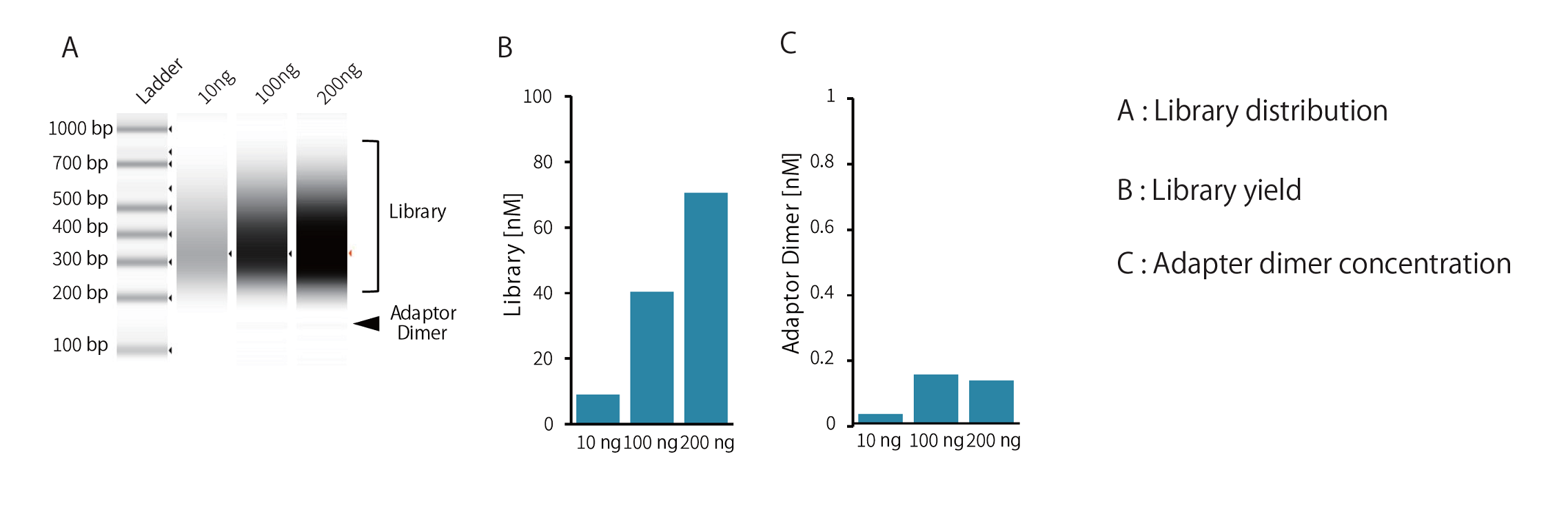
Example 4.Library preparation from various input DNA samples
Libraries were prepared from 10 ng, 100 ng, and 200 ng of ultrasonicated human genomic DNA (average fragment length: approximately 200 bp), using the GenNext™ NGS Library Prep Kit -KODEasy™-. Library distribution, yield, and adapter dimer concentration were compared, PCR amplification was performed using 5 cycles.
All genomic DNA input amounts produced yields exceeding 4 nM, which is sufficient for sequencing on Illumina platforms.
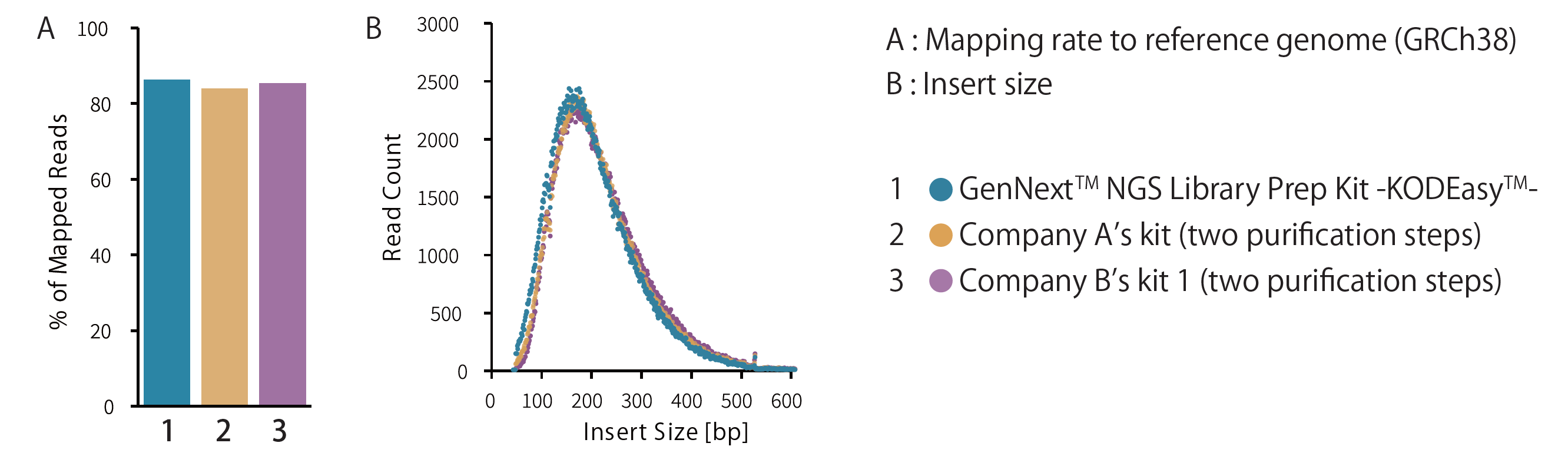
Example 5.Comparison of mapping rate and insert size by NGS
Libraries were prepared from 10 ng of ultrasonicated human genomic DNA fragmented (average fragment length: approximately 200 bp), using the GenNext™ NGS Library Prep Kit -KODEasy™-. Mapping rates and insert sizes were compared. PCR amplification was performed using 8 cycles, and sequencing was conducted with MiSeq (Illumina).
Mapping rates and insert sizes were comparable to those achieved using competitor kits with additional purification steps.
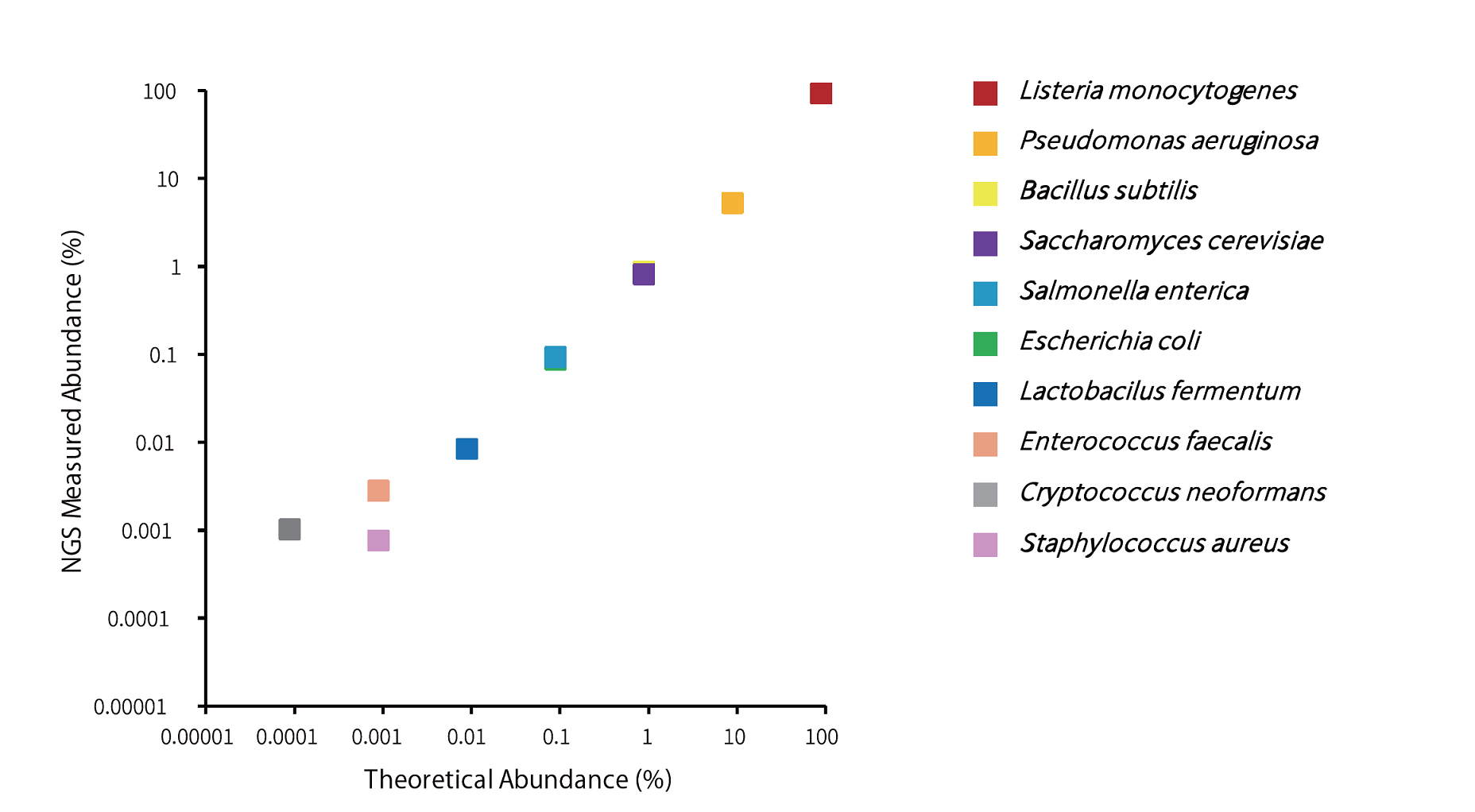
Example 6.Sequencing of mixed microbial genomic DNA
Library preparation was performed using 10ng of mixed microbial genomic DNA (ZymoBIOMICS Microbial Community DNA Standard Ⅱ (Log Distribution), Zymo Research) using the GenNext™ NGS Library Prep Kit -KODEasy™-, and the number of reads after mapping was counted. PCR was performed for 8 cycles, and sequencing was performed using MiSeq (Illumina).
A strong correlation with expected theoretical values was observed (R² = 0.97).